Usage¶
Database Connection¶
Assuming you’ve setup a successful connection and imported data through the
GUI, you can connect to the SQL database with the DVH_SQL class object.
This level of interaction with DVHA requires basic knowledge of SQL.
Refer to Data Dictionary for table and column names.
Below is an example SQL statement requesting a table with column headers of mrn, roi_name, and dvh_string such that the physician_roi is ‘brainstem’.
SELECT mrn, roi_name, dvh_string FROM DVHs WHERE physician_roi = 'brainstem';
The equivalent code in python:
from dvha.db.sql_connector import DVH_SQL
table = 'DVHs'
columns = 'mrn, roi_name, dvh_string'
condition = "physician_roi = 'brainstem'"
with DVH_SQL() as cnx:
mandible = cnx.query(table, columns, condition)
The with block is equivalent to:
cnx = DVH_SQL()
mandible = cnx.query(table, columns, condition)
cnx.close()
If no parameters are provided to DVH_SQL, it will automatically pick up
your Group 1 connection settings last used in the GUI. See the
dvha.db.sql_connector.DVH_SQL() documentation for custom connection
settings.
QuerySQL¶
Use dvha.db.sql_to_python.QuerySQL() if you’d like to query a table and
automatically convert the results into a python object more convenient than
a list of lists (as in DVH_SQL.query). The equivalent of the previous
example, using QuerySQL is below:
from dvha.db.sql_to_python import QuerySQL
table = 'DVHs'
columns = ['mrn', 'roi_name', 'dvh_string']
condition = "physician_roi = 'brainstem'"
data = QuerySQL(table, condition, columns=columns)
Or if you’d like to just pick up all columns:
data = QuerySQL(table, condition)
QuerySQL automatically adds properties based on the column name. So the
mrns are accessible with data.mrns, roi_name with data.roi_name, etc.
This works with DVHs, Plans, Rxs, and Beams tables.
DVH¶
Some data you may want with each of your DVHs is spread across multiple tables.
The dvha.models.dvh.DVH() object uses dvha.db.sql_to_python.QuerySQL()
to query the database, adds some pertinent data from other tables, and provides
many commonly used functions dealing with DVHs.
The equivalent of the previous example, using DVH is below:
from dvha.models.dvh import DVH
dvh = DVH(dvh_condition="physician_roi = 'brainstem'")
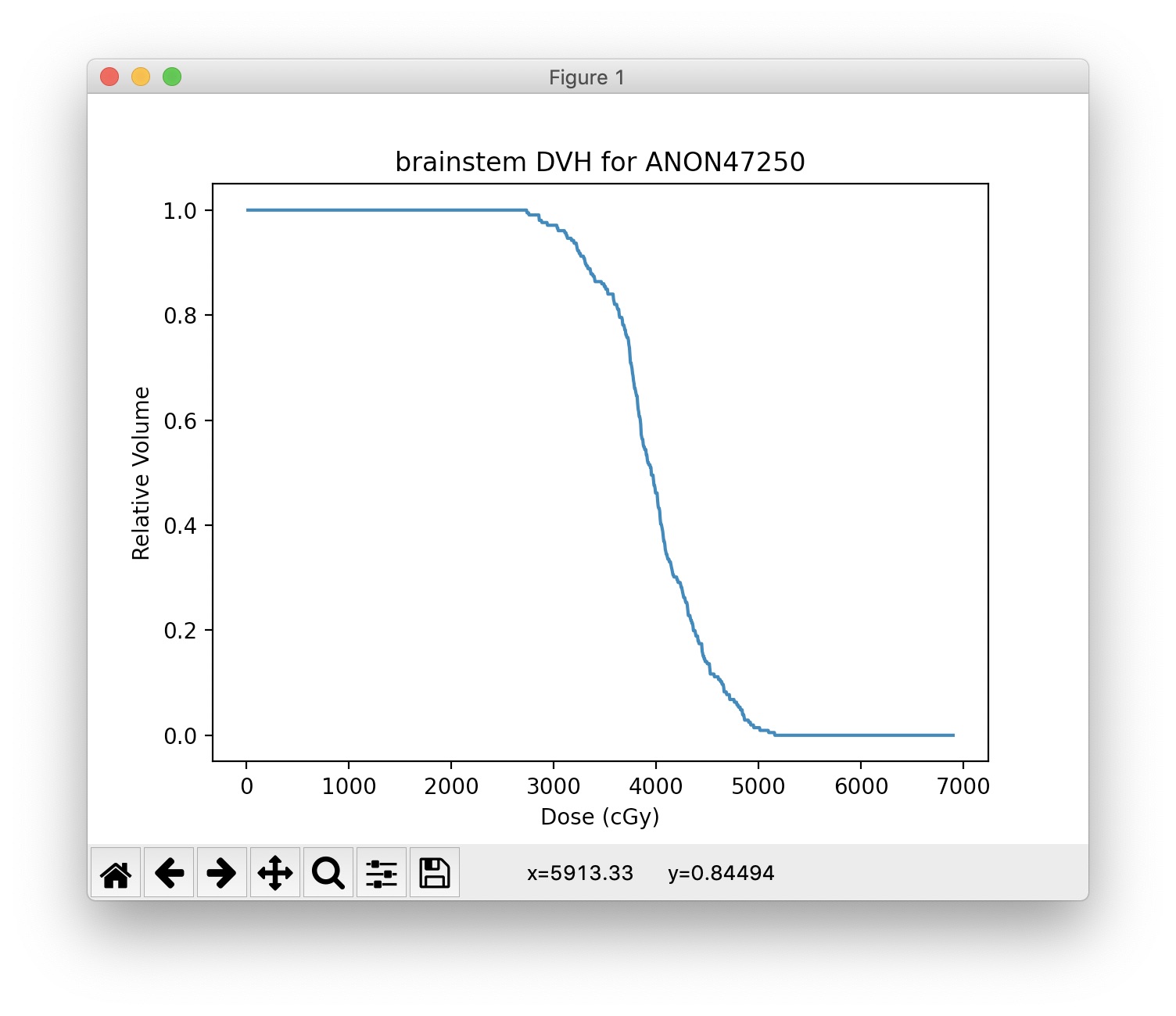
DVH Plotting¶
# Collect the plotting data
i = 0 # change this index to pick a different DVH
x = dvh.x_data[i]
y = dvh.y_data[i]
mrn = dvh.mrn[i]
roi_name = dvh.roi_name[i]
title = '%s DVH for %s' % (roi_name, mrn)
# Create the plot, may need to call plt.show() on some setups
import matplotlib.pyplot as plt
plt.plot(x, y)
plt.title(title)
plt.xlabel('Dose (cGy)')
plt.ylabel('Relative Volume')
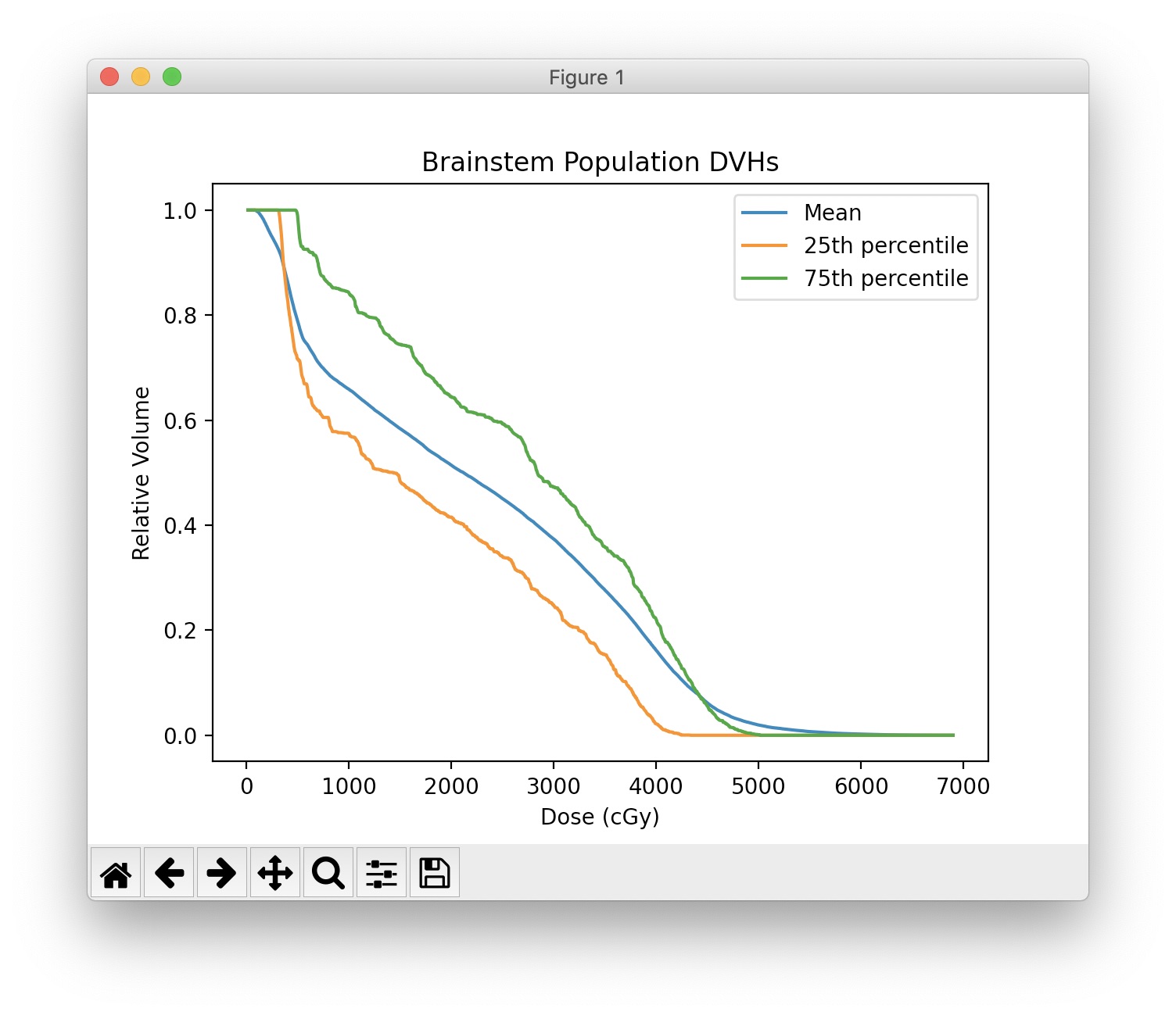
Population DVH¶
x = dvh.x_data[0]
mean = dvh.get_stat_dvh('mean')
q1 = dvh.get_percentile_dvh(25)
q3 = dvh.get_percentile_dvh(75)
plt.plot(x, mean, label='Mean')
plt.plot(x, q1, label='25th percentile')
plt.plot(x, q3, label='75th percentile')
plt.title('Brainstem Population DVHs')
plt.xlabel('Dose (cGy)')
plt.ylabel('Relative Volume')
plt.legend()
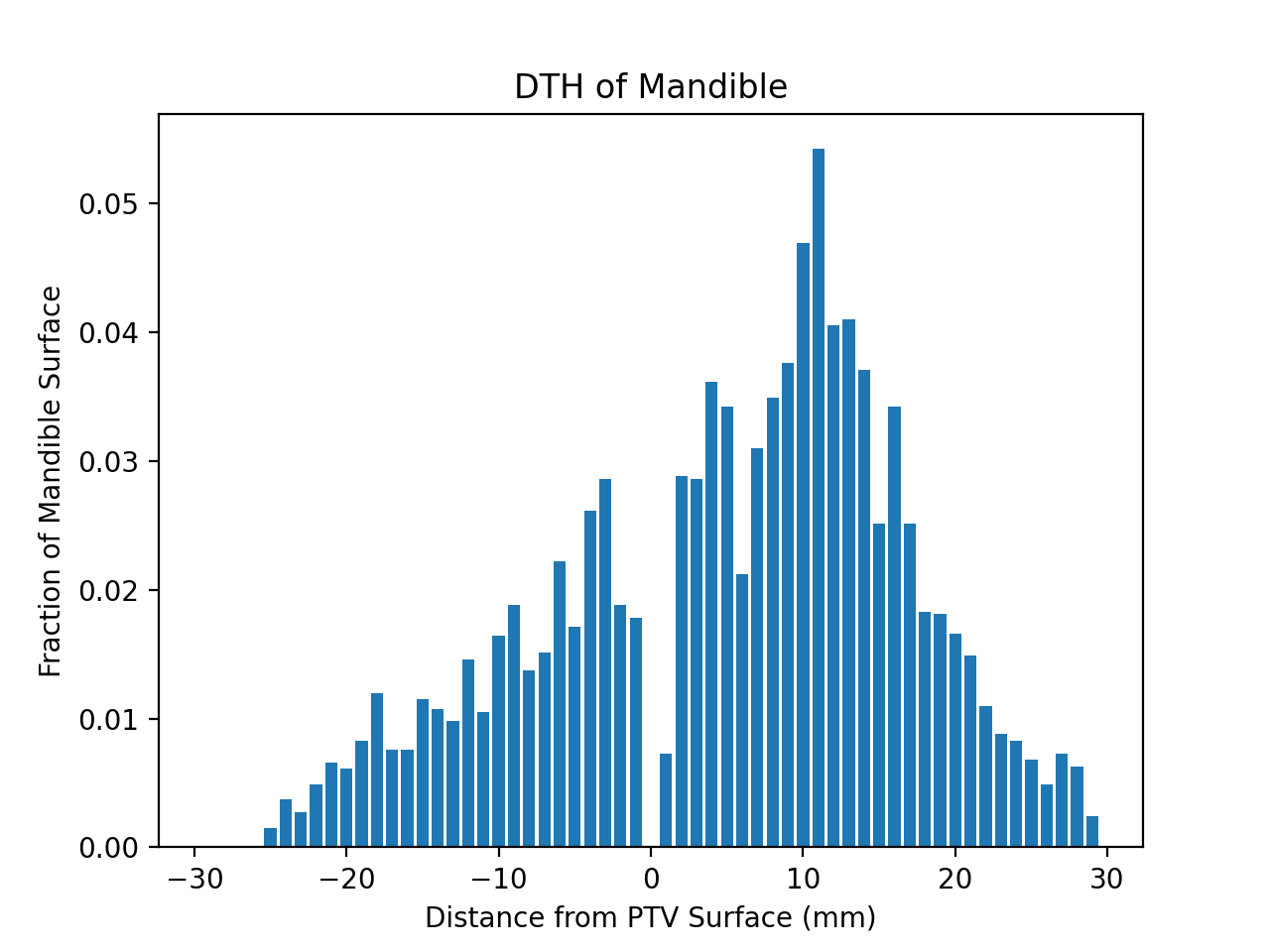
DTH¶
Although not accessible in the GUI or DVHA session data, DTHs can be extracted from the database as shown below.
from dvha.db.sql_connector import DVH_SQL
import matplotlib.pyplot as plt
from dvha.tools.roi_geometry import process_dth_string
with DVH_SQL() as cnx:
condition = "mrn = 'ANON11264' and physician_roi = 'mandible'"
mandible = cnx.query('DVHs', 'dth_string', condition)[0][0]
bins, counts = process_dth_string(mandible)
figure, axes = plt.subplots()
axes.bar(bins, counts)
axes.set_title('DTH of Mandible')
axes.set_xlabel('Distance from PTV Surface (mm)')
axes.set_ylabel('Fraction of Mandible Surface')